|
Reference:
Huang, Y.-T.,
Chao, K.-M., and
Chen, T. An Approximation
Algorithm for
Haplotype Inference by Maximum Parsimony,
Journal of Computational Biology, 12:
1261-1274, 2005.
|
Given a set of genotypes, we wish to find a minimum
subset of haplotypes that can resolve these genotypes. This problem has
been shown to be NP-hard.
 For short genotypes, we design and implement a software
called SDPHapInfer. For short genotypes, we design and implement a software
called SDPHapInfer.
|
|
 For long
genotypes, we design and implement a software called
LongHapInfer. For long
genotypes, we design and implement a software called
LongHapInfer. |
Note that the zip files are compressed using WinZip. If you
encounter any problem, please contact Prof. Kun-Mao
Chao.
|
|
SDPHapInfer
SDPHapInfer is implemented in MatLab.
The SDPHapInfer formulates the haplotype inference problem as the following
integer quadratic programming (IQP) problem. The objective function is to
minimize the number of haplotypes, and the constraint functions are to
resolve each genotype.
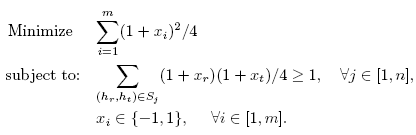
To solve the IQP problem, we propose
an iterative SDP-relaxation algorithm which finds a solution within a factor
of O(log n) of the optimal solution. The SDP problem is solved
via an external MatLab library called
SDPA-M.
|
 Download SDPHapInfer.
This zip file contains four MatLab scripts: SDPHapInfer.m, ReadGenoFile.m,
Invert.m, Chk_Resolve.m, and Find_Index.m. Download SDPHapInfer.
This zip file contains four MatLab scripts: SDPHapInfer.m, ReadGenoFile.m,
Invert.m, Chk_Resolve.m, and Find_Index.m. |
|
 Input file format: Input file format:
First line: #_of_genotypes #_of_SNPs. Second line: first genotype; Third
line: second genotype; ...
For each locus on
the genotype, we represent the homozygous wild type as 0, homozygous
mutant as 1, and heterozygous site as 2. The following is an example containing 3 genotypes with 7 SNPs.
3 7
1022101
1001222
2211002 |
|
 To run this program, type
"SDPHapInfer" under the MatLab command line, and you will see a prompt for
input of the file name. To run this program, type
"SDPHapInfer" under the MatLab command line, and you will see a prompt for
input of the file name.
>>SDPHapInfer
****************************************
*********** SDPHapInfer v0.4 ***********
****************************************
Please specify the input file name: test.dat
... |
|
 The SDPHapInfer will generate an
output file *.out (e.g., test.dat.out) that contains solutions. The SDPHapInfer will generate an
output file *.out (e.g., test.dat.out) that contains solutions.
2211002 = 0011000 + 1111001 ;
1022101 = 1001101 + 1010101 ;
1001222 = 1001101 + 1001010 ;
Number of iterations: 1
Number of selected haplotypes: 5
List of selected haplotypes:
0011000
1001010
1001101
1010101
1111001 |
|
|
LongHapInfer
LongHapInfer is implemented in MatLab. This algorithm partitions the
genotypes into segments, infers subhaplotypes for each segment, and
concatenates subhaplotypes in different segments for the final solution.
LongHapInfer requires an external MatLab library called
SDPA-M.
|
 Download
LongHapInfer. This zip file contains four MatLab scripts:
LongHapInfer.m, ReadGenoFile.m, Invert.m, Chk_Resolve.m, and Find_Index.m. Download
LongHapInfer. This zip file contains four MatLab scripts:
LongHapInfer.m, ReadGenoFile.m, Invert.m, Chk_Resolve.m, and Find_Index.m. |
|
 Input file format: Input file format:
First line: #_of_genotypes #_of_SNPs. Second line: first genotype; Third
line: second genotype; ...
For each locus on
the genotype, we represent the homozygous wild type as 0, homozygous
mutant as 1, and heterozygous site as 2. The following is an example containing 3 genotypes with 7 SNPs.
3 7
1022101
1001222
2211002 |
|
 To run this program, type
"LongHapInfer" under the MatLab command line, and you will see a prompt
for
input of the file name. To run this program, type
"LongHapInfer" under the MatLab command line, and you will see a prompt
for
input of the file name.
>>LongHapInfer
****************************************
*********** LongHapInfer v0.1 ***********
****************************************
Please specify the input file name: test.dat
... |
|
 The LongHapInfer will generate an
output file *.out (e.g., test.dat.out) that contains solutions. The LongHapInfer will generate an
output file *.out (e.g., test.dat.out) that contains solutions. |
|